New paper: Unconstraining graph-constrained group testing
We just submitted a new paper on Graph-Constrained Group Testing which I’m really excited about!
The problem we consider is related to debugging networks: suppose you have a network where some of the links in your network fail. You can send a packet around the network, and observe whether it reaches its destination or not. How many packets do you need to send to find the failing links?
This is my blog, so here’s a bonus version of the problem. Suppose you manage a large national park with lots of hiking trails. Last night there was a huge storm, and a few trails (at most $d$) have been washed out. You would like to know which trails were washed out before you reopen the park, so that you can warn visitors. The catch is that in this universe, the only way you can tell if a trail has been washed out is to hire a hiker and send them on some walk around the park. If the hiker comes back, you know every trail they visited was good. However, if they find a washed out trail, they’ll lose the trail and wander around the woods for a bit, so you know that some trail they visited was bad. You could send out hikers one by one and wait for them to get back, but hey, you have a park to open here! The question is: what is the minimum number of hikers you need to send out simultaneously to find all the washed out trails?
Here’s the more formal version of this problem. You have a graph $G = (V,E)$ where $|V| = n, |E| = m$ and some set $B \subseteq E$ of the edges are defective. $B$ is a small set, of size at most $d$. You can test some connected subgraph $G’$ of $G$ and determine whether any edge in $B$ is in $G’$. The problem is to find the minimum number of non-adaptive tests such that $B$ can be exactly identified from the test results.
If there were no graph in this problem, this would be a classic problem called Combinatorial Group Testing. During World War II, the military was interested in finding soldiers with syphilis using some blood test. Syphilis tests were very expensive, and the military didn’t want to test everyone individually. In a nice paper from 1948, Dorfman showed that by testing groups (i.e. mix a bunch of blood in a bucket and test the bucket), one could find the sick soldiers using many fewer tests.
Group testing (sans graph constraints) can be solved using roughly $O(d^2 \log m)$ tests. With the graph constraints, the answer is less well-known. [Harvey 2007] gave the answer for a number of special cases, and showed that roughly $O(d^3 \log m)$ tests were enough for certain graphs. In a later work, [Cheraghchi 2010] showed a cool algorithm using random walks which uses roughly $O(d^2 \text{polylog } m)$ tests for good expanders and, surprisingly, $O(d^2 \log m)$ tests for a complete graph. That is, group testing with the constraints added by the complete graph is in some sense no more difficult than group testing without constraints.
We show the following result: for many graphs, you can do group testing in only $O(d^2 \log m)$ tests! As long as the graph is sufficiently well-connected, you can do group testing with graph constraints “for free.” The graphs this works for includes complete graphs, good expanders (e.g. random regular graphs and Erdos-Renyi graphs), and graphs which are “clusters” of good expanders (e.g. a barbell).
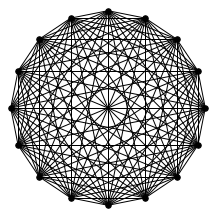
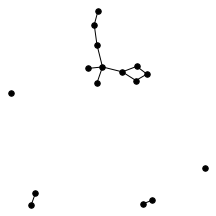
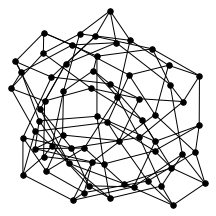
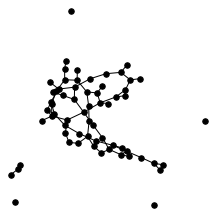
There’s also a cool technical result about “giant components” of graphs. Suppose you have a graph, and sparsify it by constructing a subgraph which includes each edge independently with probability $p$. What does this subgraph look like? For the complete graph, this question was studied by Erdős and Rényi in the 50s, and it turns out that a “giant component” - a connected component which includes lots of nodes - appears at a certain threshold. For $p=\frac{1-\epsilon}{n}$, the largest connected component has size $O(\log n)$. For $p = \frac{1+\epsilon}{n}$, the largest has size $O(n)$. It turns out (and has been known for a while) the same thing happens in expander graphs. We prove a slightly stronger version of this result about expanders, which was helpful in analyzing our algorithm. You can check out the paper for the full details, but for now let me just show some neat pictures of giant components: